An Example Ecosystem Workflow
Ecosystem Workflow — Data, Compute and Visualisation from the Office, Home and Abroad
By using the ecosystem computational facilities, storage and virtual desktop, it is easy to prepare, submit and monitor jobs, and post-process results, without transferring any data, all from home, office and abroad (e.g. while at a conference).
Example Workflow
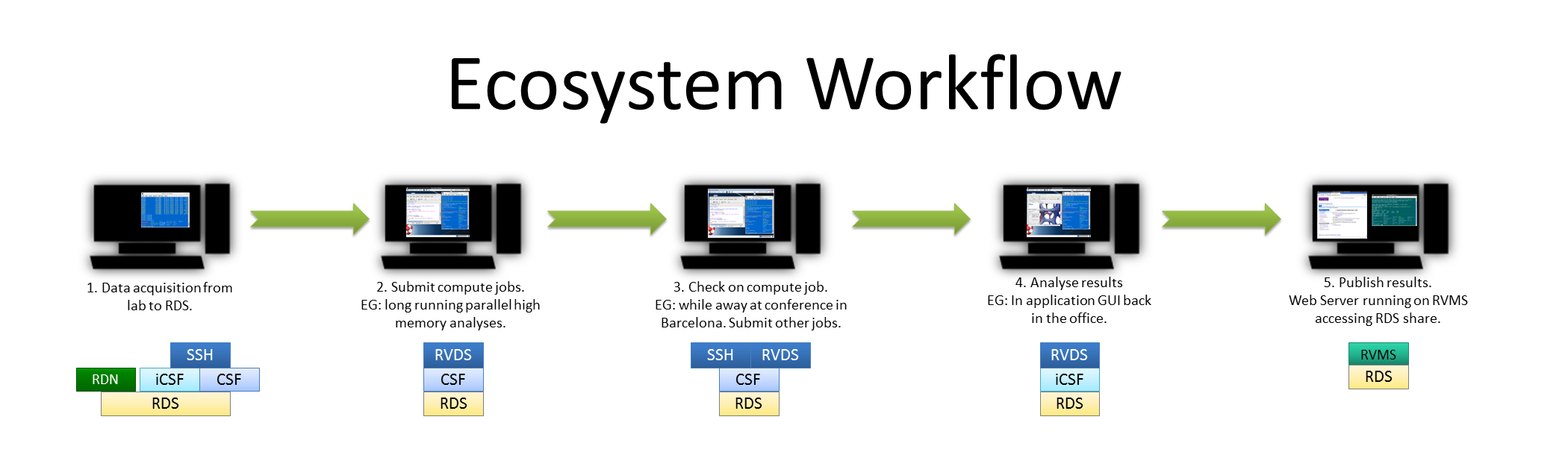 CIR Example Workflow in Life Sciences (click for larger image.)
CIR Example Workflow in Life Sciences (click for larger image.)- A research’s data exits their group’s DNA sequencer straight onto storage provided by the RDS, over fast, dedicated networking infrastructure (the RDN). The data is now visible on the CSF and iCSF.
- The researcher then launches a session on the RVDS. From this desktop, they access the CSF to define a series of computational jobs to process the data – these are submitted to the batch system.
- Later, from home, the researcher re-connects to the RVDS session to monitor the jobs to ensure all is well — or make any necessary tweaks. Over the next few days, from a conference in Barcelona, the researcher can check progress again using the RVDS, from their laptop – for example, they clear some jobs which have failed and submit additional, corrected work.
- Once back at the office, the batch jobs have finished, so using the same RVDS session, the researcher starts a GUI-based, interactive post-processing app on Incline (aka the iCSF) — no need to move data as all the same RDS-based filesystems are available on all ecosystem compute resources.
- Finally, the results are ready and made available to the public via a Web server running on the Research Virtual Machine Service (RVMS) — accessing the same RDS share.
